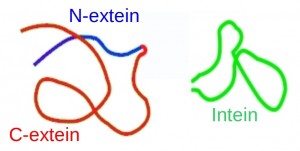
 Many proteins are synthesized in biologically inactive forms, and are activated in post-translational processes such as proteolytic cleavage. This process is usually catalyzed by external proteins, but some proteins are able to self-catalyze the reactions without the need for any other protein or cofactor. We are interested in one of this post-translational process, known as protein splicing. In protein splicing, a segment of an inactive protein, the intein (internal protein), is excised from the rest of the protein, and the two flanking domains, the C- and N-exteins, join each other, forming a biologically active protein (see Figure).
Many proteins are synthesized in biologically inactive forms, and are activated in post-translational processes such as proteolytic cleavage. This process is usually catalyzed by external proteins, but some proteins are able to self-catalyze the reactions without the need for any other protein or cofactor. We are interested in one of this post-translational process, known as protein splicing. In protein splicing, a segment of an inactive protein, the intein (internal protein), is excised from the rest of the protein, and the two flanking domains, the C- and N-exteins, join each other, forming a biologically active protein (see Figure).
From a biochemical point of view the process is very interesting because In vitro observations,8,9 between two foreign proteins, have demonstrated that only amino acids located in the intein are directly involved in the process, and that it requires no other proteins or external sources of metabolic energy such as ATP. Therefore, the scission of the intein from the rest of the protein is completely self-catalyzed by the intein itself. This weird feature of protein splicing has facilitated its use as a biotechnological tool and its application in protein engineering, chemical biology or medicine.
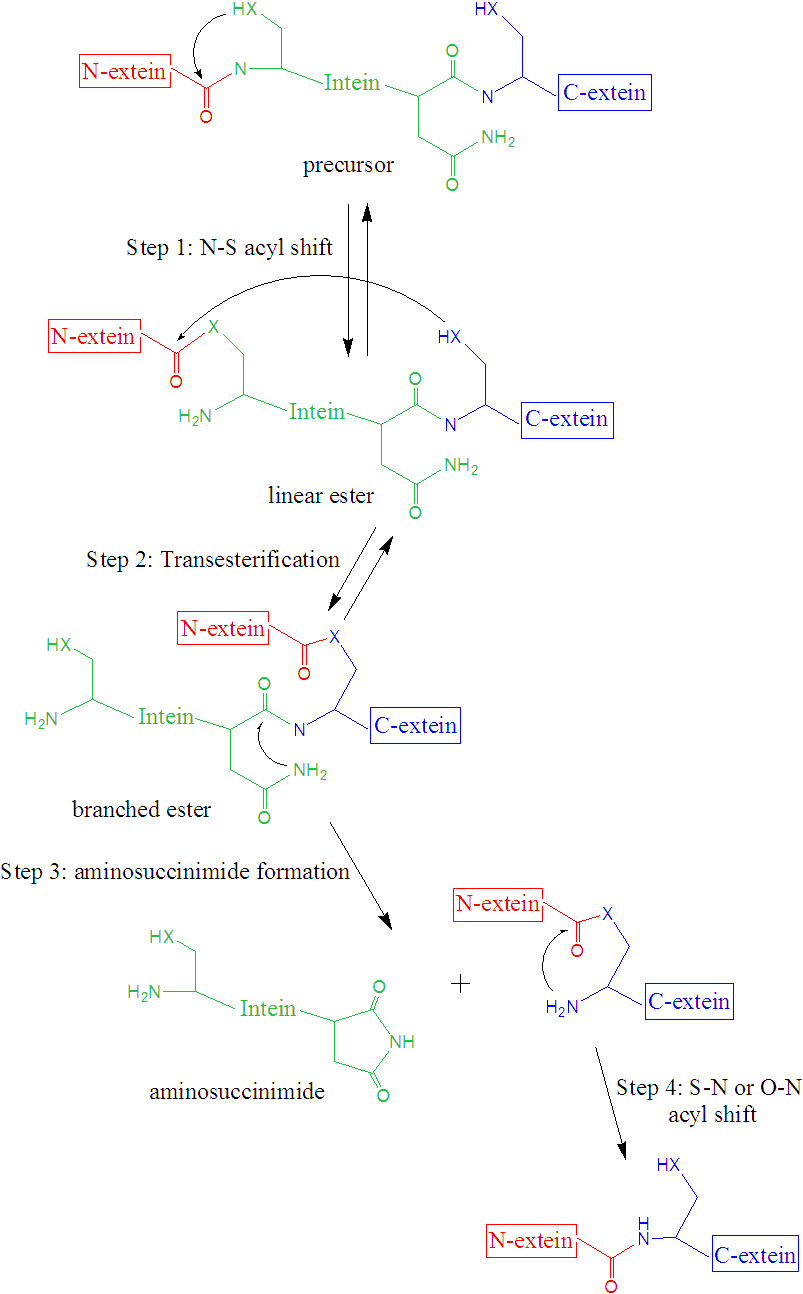
Even that the overall mechanism of the process is well established, the mechanistic details about how the process takes place remains to be fully understood. The overall mechanism is usually (there are some exceptions) formed by four steps: i) N-O or N-S acyl rearrangement to form a linear ester intermediate, ii) transesterification to form a branched ester intermediate, iii) cyclization of an Asn (or Gln), coupled with the cleavage of the intein-C-extein peptide bond, forming an aminosuccinimide residue and an ester, and iv) spontaneous hydrolysis of the aminosuccinimide residue and S-N or O-N rearrangement of the ester between the exteins to form a more stable amide (peptide) bond.
Our group has paid attention on the first and third steps, since a peptide bond is cleaved in them. We have combined different techniques to tackle the study, in which the intein was isolated and inserted such as highly-accurate quantum chemical methods, molecular dynamics simulations and high level QM/MM hybrid calculations.
Publications of the group in the area:
- Mujika J.I., Lopez X. “Protein Splicing” Encyclopedia of Biophysics, 2013, DOI 10.1007/978-3-642-16712-6
- Mujika J.I., Lopez X., Mulholland A. J. “Mechanism of C-terminal intein cleavage in protein splicing from QM/MM molecular dynamics simulations” Org. Biomol. Chem., 2012, 10, p1207-1218
- Mujika J.I., Lopez X., Mulholland A. J. “Modeling Protein Splicing: Reaction Pathway for C-Terminal Splice and Intein Scission” J. Phys. Chem. B, 2009, 113, p5607-5616